-Search query
-Search result
Showing all 45 items for (author: da & fonseca & p)

EMDB-40758: 
Isobutyryl-CoA mutase fused in the presence of GMPPCP
Method: single particle / : Vaccaro FA, Drennan CL

PDB-8sta: 
Isobutyryl-CoA mutase fused in the presence of GMPPCP
Method: single particle / : Vaccaro FA, Drennan CL

EMDB-23355: 
Structure of dNTPase at 3.3 Angstrom Resolution
Method: single particle / : Bouvette J, Liu HF, Du X, Zhou Y, Sikkema AP, Mello JFR, Klemm B, Huang R, Schaaper RM, Borgnia MJ, Bartesaghi A

EMDB-23356: 
Structure of dNTPase at 3.6 Angstrom Resolution
Method: subtomogram averaging / : Bouvette J, Liu HF, Du X, Zhou Y, Sikkema AP, Mello JFR, Klemm B, Huang R, Schaaper RM, Borgnia MJ, Bartesaghi A

EMDB-23357: 
Structure of 80S ribosome from EMPIAR-10064 at 5.6 Angstrom Resolution
Method: subtomogram averaging / : Bouvette J, Liu HF, Du X, Zhou Y, Sikkema AP, Mello JFR, Klemm B, Huang R, Schaaper RM, Borgnia MJ, Bartesaghi A

EMDB-23358: 
Structure of E. coli 70S ribosome from EMPIAR-10304 at 4.8 Angstrom Resolution
Method: subtomogram averaging / : Bouvette J, Liu HF, Du X, Zhou Y, Sikkema AP, Mello JFR, Klemm B, Huang R, Schaaper RM, Borgnia MJ, Bartesaghi A

EMDB-11328: 
SARS-CoV-2 spike in prefusion state
Method: single particle / : Martinez M, Marabini R, Carazo JM

EMDB-11336: 
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation)
Method: single particle / : Martinez M, Marabini R, Carazo JM

EMDB-11337: 
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation)
Method: single particle / : Martinez M, Marabini R, Carazo JM

EMDB-11341: 
SARS-CoV-2 stabilized spike in prefusion state (1-up conformation)
Method: single particle / : Martinez M, Marabini R, Carazo JM

PDB-6zow: 
SARS-CoV-2 spike in prefusion state
Method: single particle / : Martinez M, Marabini R, Carazo JM

PDB-6zp5: 
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation)
Method: single particle / : Martinez M, Marabini R, Carazo JM

PDB-6zp7: 
SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation)
Method: single particle / : Martinez M, Marabini R, Carazo JM

EMDB-4860: 
Human 20S-PA200 Proteasome Complex
Method: single particle / : Toste Rego A, da Fonseca PCA

EMDB-4877: 
Human 20S Proteasome
Method: single particle / : Toste Rego A, da Fonseca PCA

PDB-6rey: 
Human 20S-PA200 Proteasome Complex
Method: single particle / : Toste Rego A, da Fonseca PCA

PDB-6rgq: 
Human 20S Proteasome
Method: single particle / : Toste Rego A, da Fonseca PCA

EMDB-0347: 
Apo form metabotropic glutamate receptor 5 with Nanobody 43
Method: single particle / : Koehl A, Hu H, Feng D, Zhang Y, Sun B, Kobilka TS, Pardon E, Steyaert J, Mathiesen JM, Skiniotis G, Kobilka BK

EMDB-0345: 
Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43
Method: single particle / : Koehl A, Hu H, Feng D, Sun B, Weis WI, Skiniotis GS, Mathiesen JM, Kobilka BK

EMDB-0346: 
Metabotropic Glutamate Receptor 5 Apo Form
Method: single particle / : Koehl A, Hu H, Feng D, Sun B, Weis WI, Skiniotis GS, Mathiesen JM, Kobilka BK

PDB-6n51: 
Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43
Method: single particle / : Koehl A, Hu H, Feng D, Sun B, Weis WI, Skiniotis GS, Mathiesen JM, Kobilka BK

PDB-6n52: 
Metabotropic Glutamate Receptor 5 Apo Form
Method: single particle / : Koehl A, Hu H, Feng D, Sun B, Weis WI, Skiniotis GS, Mathiesen JM, Kobilka BK

PDB-5fmg: 
Structure and function based design of Plasmodium-selective proteasome inhibitors
Method: single particle / : Li H, O'Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PCA, Bogyo M

EMDB-3231: 
Structure and function based design of Plasmodium-selective proteasome inhibitors
Method: single particle / : Li H, O' Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PCA, Bogyo M

PDB-5a0q: 
Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core
Method: single particle / : daFonseca PCA, Morris EP

EMDB-2981: 
Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core
Method: single particle / : da Fonseca PCA, Morris EP

EMDB-2226: 
Single particle analysis of recombinant human anaphase promoting complex (APC/C)
Method: single particle / : Zhang Z, Yang J, Kong EH, Chao WCH, Morris EP, da Fonseca PCA, Barford D
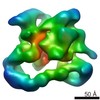
EMDB-2173: 
Negative stain electron microscopy of a CSN-SCF~Nedd8/Skp2/Cks1 complex
Method: single particle / : Enchev RI, Scott DC, da Fonseca P, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP
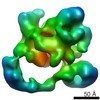
EMDB-2174: 
Negative stain electron microscopy of a CSN-SCF~Nedd8/Fbw7 complex
Method: single particle / : Enchev RI, Scott DC, da Fonseca P, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP
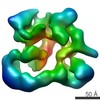
EMDB-2175: 
Negative stain electron microscopy of a CSN(dCsn5)-SCF~Nedd8/Skp2/Cks1 complex
Method: single particle / : Enchev RI, Scott DC, da Fonseca P, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP

EMDB-2176: 
Negative stain electron microscopy of a CSN complex
Method: single particle / : Enchev RI, Scott DC, da Fonseca P, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP

EMDB-2177: 
Negative stain electron microscopy of a CSN complex
Method: single particle / : Enchev RI, Scott DC, da Fonseca P, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP

EMDB-2047: 
Structure of the human 26S proteasome
Method: single particle / : da Fonseca PCA, He J, Morris EP

EMDB-2026: 
Structure of the proteasome subunit Rpn1
Method: single particle / : He J, Kulkarni K, daFonseca P, Krutauz D, Glickman M, Barford D, Morris E

EMDB-1842: 
Structural basis for the subunit assembly of the anaphase promoting complex
Method: single particle / : Schreiber A, Stengel F, Zhang Z, Enchev RE, Kong E, Morris EP, Robinson CV, daFonseca PCA, Barford D
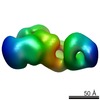
EMDB-1843: 
Structural basis for the subunit assembly of the anaphase promoting complex
Method: single particle / : Schreiber A, Stengel F, Zhang Z, Enchev RE, Kong E, Morris EP, Robinson CV, daFonseca PCA, Barford D

EMDB-1841: 
Structural basis for the subunit assembly of the anaphase promoting complex
Method: single particle / : Schreiber A, Stengel F, Zhang Z, Enchev RE, Kong E, Morris EP, Robinson CV, daFonseca PCA, Barford D

EMDB-1844: 
Structural basis for the subunit assembly of the anaphase promoting complex
Method: single particle / : Schreiber A, Stengel F, Zhang Z, Enchev RE, Kong E, Morris EP, Robinson CV, daFonseca PCA, Barford D

EMDB-1845: 
Structural basis for the subunit assembly of the anaphase promoting complex
Method: single particle / : Schreiber A, Stengel F, Zhang Z, Enchev RE, Kong E, Morris EP, Robinson CV, daFonseca PCA, Barford D

EMDB-1815: 
The structure of anaphase promoting complex-Cdh1-Dbox at 10 Angstroms resolution
Method: single particle / : daFonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D

EMDB-1817: 
Structure of S. cerevisiae anaphase promoting complex with the co-activator Cdh1
Method: single particle / : daFonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D

EMDB-1818: 
Structure of S. cerevisiae anaphase promoting complex-Cdh1-KENbox peptide
Method: single particle / : daFonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D

EMDB-1816: 
Structure of S. cerevisiae anaphase promoting complex
Method: single particle / : daFonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D

EMDB-1819: 
Structure of S. cerevisiae anaphase promoting complex-Cdh1 bound to a Hsl1 fragment
Method: single particle / : daFonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D

EMDB-1617: 
C12 symmetrised 3D reconstruction of the Shigella flexneri T3SS needle complex from negatively stained sample electron micrographs
Method: single particle / : Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da Fonseca PCA, Morris EP, Wall JS, Lea SM, Blocker AJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
